Programmable Nanomaterials
The ability to synthesize materials by design is one of the greatest outstanding scientific challenges. Advances towards meeting this challenge will lead to discoveries in fields such as plasmonics, photonics, catalysis, and energy sciences.
In the Mirkin group, we employ a novel approach to the realization of materials by design, where nanoparticle “atoms” functionalized with a dense shell of oligonucleotides are assembled via oligonucleotide “bonds” into crystalline superlattices with tunable compositions, symmetries, and lattice parameters. The high degree of control over the assembly characteristics of these oligonucleotide-nanoparticle conjugates, termed “programmable atom equivalents” (PAEs), is enabled by the exceptional molecular recognition, tunability, and predictability of the nucleic acid bonds (Fig. 1). Unlike atomic systems in which the bonding character of an atom is dictated by its atomic composition and configuration of valence electrons, PAEs allow one to independently control the nanoparticle core (tunability in the nanoparticle size, shape, or composition) separately from the oligonucleotide “bonds” (controllable nucleic acid type, sequence, length, flexibility, and strength).
While PAEs provide discrete control over crystal engineering where the location of every nanoparticle is fixed, we discovered that small nanoparticles (~1.5 nm) functionalized with DNA can be mobile within nanoparticle lattices (Fig. 2). Specifically, these small particles, termed electron equivalents (EEs), readily diffuse within stabilize lattices of repulsive PAEs. The diffusion (i.e., delocalization) of EEs introduces the concept of metallicity to colloidal crystal engineering, where metallic, intermetallic, and other complex phases can now be targeted. Ongoing research in the Programmable Nanomaterials subgroup focuses on exploiting the functionalities of the nanoparticle atoms in the crystalline state, tuning the nature of the DNA bonds, and studying the diffusive properties of small nanoparticles functionalized with DNA.
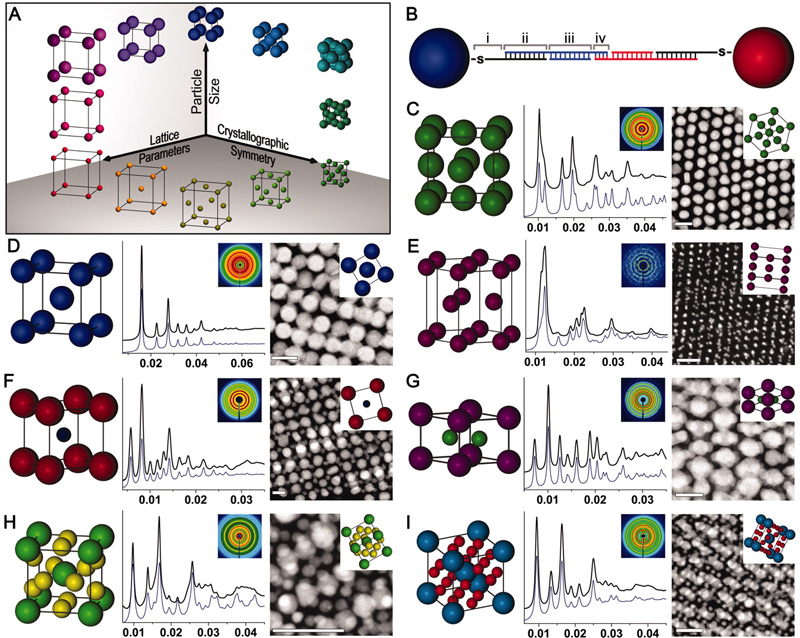
Figure 1. Nanoparticle superlattice engineering with DNA. (A) Particle size, lattice parameters, and lattice symmetry can be independently changed through a (B) sticky end-mediated DNA design. Over 70 symmetries are accessible, including (C) fcc, (D) bcc, (E) hcp, (F) CsCl, (G) AlB2, (H) Cr3Si, and (I) Cs6C60 lattices. (Adapted from Science 2011, 334, 204.)
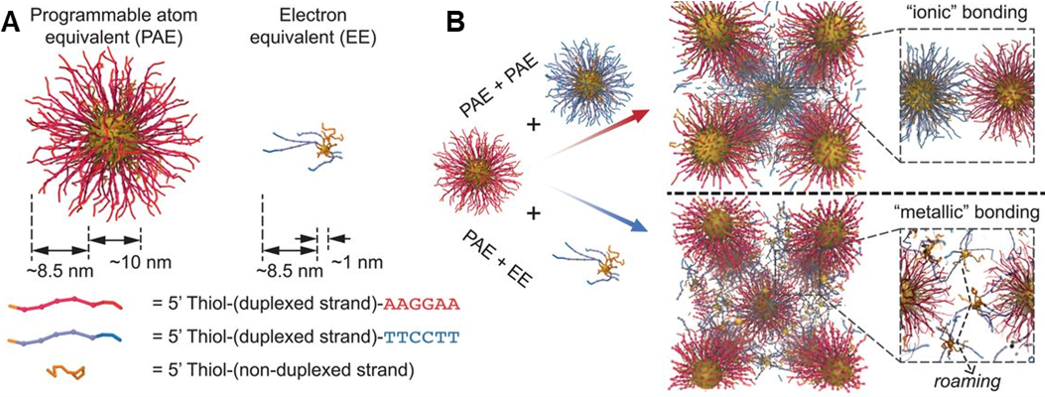
Figure 2. Colloidal crystal engineering using electron equivalents. (A) Illustration of PAEs versus EEs, where particle size and the number of DNA strands influence the properties of the DNA-functionalized nanoparticles. (B) When two types of PAEs with similar sizes and complementary DNA sequences are assembled, ionic bonding is observed in crystal lattices. In contrast, co-assembling PAEs and EEs results in metallic bonding characteristics due to the diffusive properties of EEs. (Adapted from Science 2019, 364, 1174.)
Integrating nanoparticle functionalities into superlattices
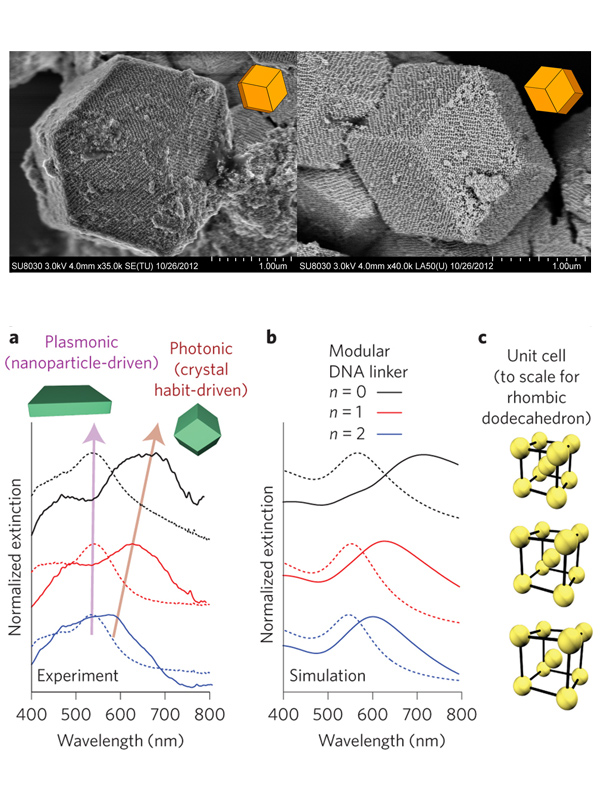
Nanoparticles have interesting catalytic and optical properties that are distinct from their atomic or molecular precursors and are often dependent on their assembly state. We are exploiting the programmability of the DNA bond to tune the structures of PAE superlattices assembled in solution or layer-by-layer on substrates and investigating how superlattice architecture influences light-matter interactions. Nanoparticle superlattices composed of Au nanoparticle-cored PAEs show strong light-plasmon interactions that were tunable by independently controlling lattice constants and nanoparticle diameters (Fig. 3). By employing a layer-by-layer assembly approach, the thickness of the crystal can be modulated to control the relative contributions of plasmonic and photonic properties to the overall light-matter interactions of the crystal. We are combining these experimental studies with a theoretical framework that will allow us to predict how crystal lattice structure, habit, and composition control light-matter interactions, and ultimately, allow us to develop highly tunable plasmonic metamaterials.
Figure 3. Structure-function relationships in superlattices. (Top) Slow cooling of PAEs produces single crystals with the expected equilbirum crystal habit. (Bottom) (a) Experimental and (b) simulated extinction spectra for thin-films and superlattices constructed with different DNA linkers. (c) Scale rendering of the unit cells for the lattices measured in (a). (Adapted from (top) Nature 2014, 505, 73 and (bottom) Nat. Nanotechnol. 2015, 10, 453.)
Utilizing the chemical addressability of proteins for DNA-directed assembly
Proteins are archetypal nanoscale building blocks that assemble into functional materials with applications including catalysis, energy transfer, and structural actuation. However, programming the interactions between proteins, and their positions within materials, is challenging due to the various interactions which typify protein-protein interfaces. The Mirkin group is taking advantage of the diverse structures and the inherent or introduced orthogonal surface chemistries of proteins to control their assembly (Fig. 4) and develop polymeric protein systems, protein single crystals, and multicomponent superlattices with symmetries that are difficult to obtain using spherical PAEs. Ultimately, these protein-based materials will be designed to integrate the intrinsic functionalities of each protein building block selected into programmable hybrid materials (e.g., with inorganic nanoparticles).
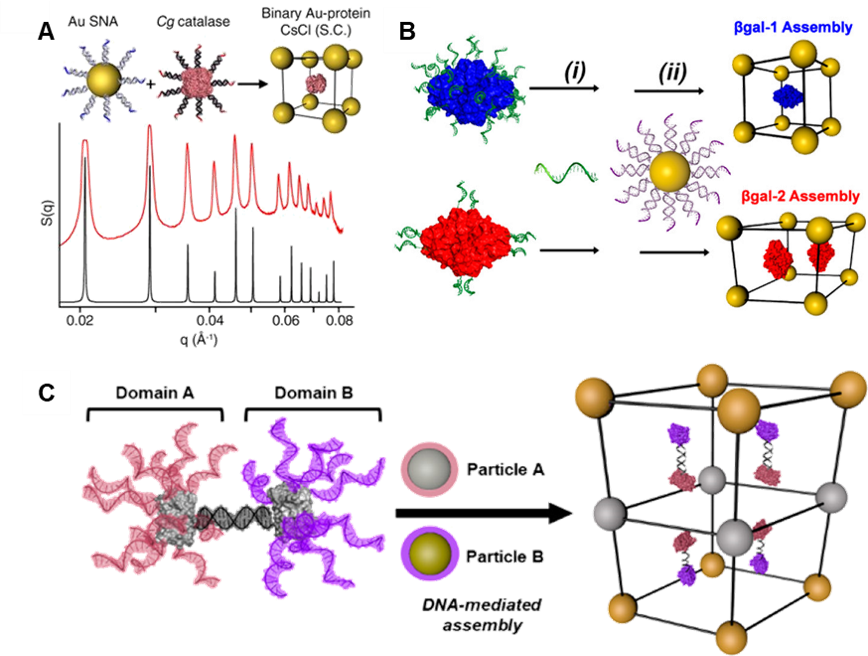
Figure 4. Protein material engineering with DNA. (A) Dense functionalization of proteins with DNA produces protein PAEs that crystallize with AuNPs. (B) Tuning the distribution of DNA on the protein surface directs assembly outcome. (C) Site-specific Janus protein PAEs direct inorganic nanoparticles to assemble into otherwise inaccessible layered structures. (Adapted from (A) Proc. Natl. Aca. Sci. USA 2015, 112, 4564; (B) J. Am. Chem. Soc. 2017, 139, 1754; and (C) J. Am. Chem. Soc. 2018, 140, 9269.)
Employing the programmability of the DNA bond to create tunable and responsive nanoparticle superlattices
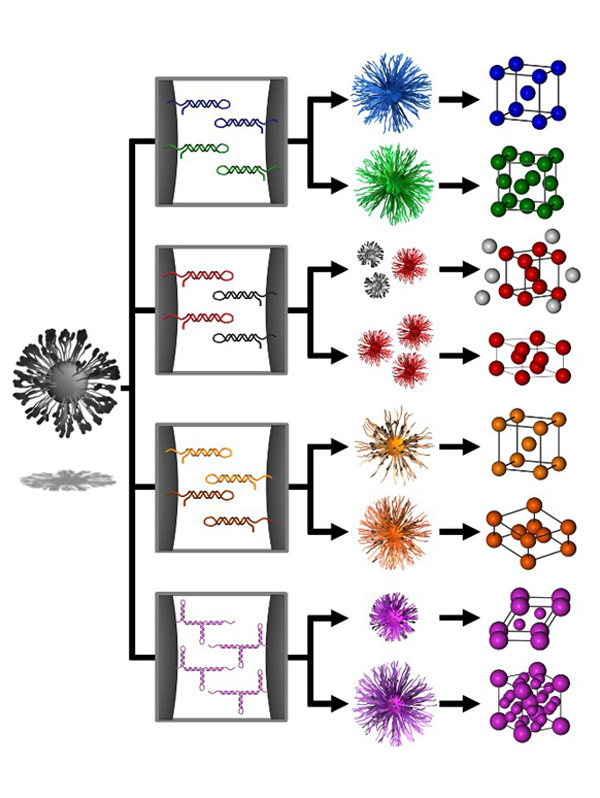
Biological systems are inherently responsive to external stimuli, enabling a high degree of control over complex processes such as signaling cascades. We are developing several unique approaches for designing oligonucleotide bonds that enable a similar degree of responsiveness and tunability to be incorporated into PAE superlattices. One example of this is the concept of “transmutable nanoparticles,” which include a number of closed ‘hairpin’ DNA strands along the surface of nanoparticles that cannot link with strands on neighboring particles (Fig. 5). The addition of “effector” strands, which are short oligonucleotide sequences that act as switches and can open these hairpins, allows the particles to bond into a particular lattice arrangement. By using different switches and combinations of nanoparticles, we can effectively and dynamically change the bonding character of “transmutable nanoparticles” and drive their crystallization pathway along multiple thermodynamic trajectories.
Figure 5. Transmutable superlattices via reconfigurable DNA ligands. DNA hairpins act as unactivated DNA ligands that can be selectively activated in the presence of an effector strand. Selective deprotection of PAEs with self-complementary or non-self-complementary strands leads to distinct structural outcomes. (Adapted from Science 2016, 351, 579.)
We have also tuned the stability and responsiveness of PAE superlattices through multiple other means (Fig. 6), including the replacement of DNA-DNA bonds with RNA-RNA or RNA-DNA bonds; the stabilization of superlattices with multivalent metal cations; and the incorporation of light- and pH-responsive moieties into DNA bonds. These different types of oligonucleotide bonds result in superlattices with unique properties that affect their thermal stability, susceptibility to enzymatic degradation, or light responsiveness, for example.
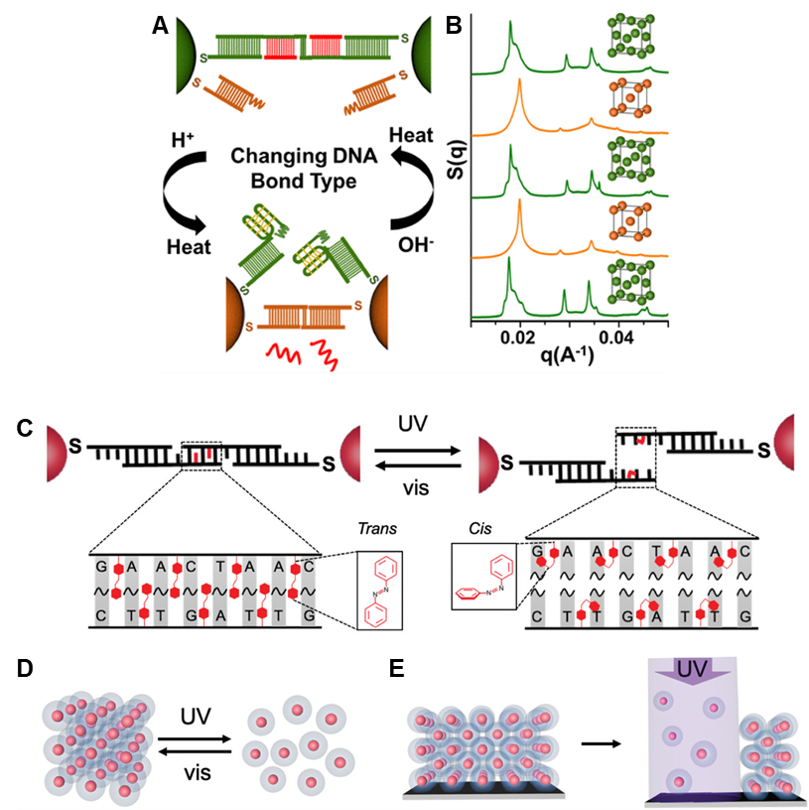
Figure 6. Reconfigurable, stimuli-responsive superlattices. (A) Nanoparticles with i-motif-containing self-complementary sticky ends and non-i-motif-containing complementary sticky ends can be switched between bonding types in response to pH change, leading to (B) lattice symmetry switching between fcc and bcc. (C) Azobenzene-containing DNA ligands lead to (D) light-responsive assembly and disassembly. (E) Irradiation of a surface-bound thin film selectively disassembles specific areas of the superlattice. (Adapted from (A,B) Adv. Mater. 2020, 32, 1906600 and (C,D,E) J. Am. Chem. Soc. 2018, 140, 5061.)
Programmable Materials Subgroup Members

Back row (L-R): Vinzenz Mayer, Taokun Luo, Jacob Cohen, John Cavaliere
Middle row (L-R): Xiaowei Liu, Jun Li, T. Ocampo, Maya Kesan, Jack Ahrens, , Allen Guo, Rachel Chan, Mia Pascall, Tanner Fink (subgroup leader)
Front Row (L-R): Namrata Ramani (Subgroup leader), Ana Leal, Jennifer Delgado, Hadley McCormick, Yinglin Ma, Alex Cushing, Janice Kang, Julianna Burgeois