Nanomedicine
Research within this subgroup focuses on synthesizing and applying spherical nucleic acids (SNAs) for the treatment of disease, gaining an understanding of how nanomaterials interact with other entities in complex biological environments, establishing structure-activity relationships for nanomaterials in biological responses, and understanding the mechanisms of SNA trafficking within cells and tissues. Ultimately, we seek to explore the range of therapeutic applications for SNAs that exploit these important observations and insights.
What are SNAs?
Spherical nucleic acids, or SNAs, (Fig. 1) are three-dimensional nanostructures, consisting of nucleic acids that are densely functionalized and oriented spherically around a nanoparticle core (like a “koosh ball”). These constructs define a new class of nucleic acids that have properties that markedly differ from those of the analogous, linear (1D) strands of the same sequence. For example, on a sequence-for-sequence basis, SNAs bind complementary nucleic acids tighter (orders of magnitude) and enter cells more rapidly and in higher quantities without the use of transfection agents by engaging scavenger receptors that facilitate caveolin-mediated endocytosis. These abilities have enabled the use of SNAs in biological applications, spanning extra- and intracellular detection, gene regulation, and immunotherapy (see Video 1). SNAs also are fundamental building blocks in material science – a type of “programmable atom equivalent” (PAE) – and have paved the way for the introduction of a new form of chemistry based on nanoparticles as “atoms” and DNA strands as programmable “bonding” elements. PAEs, through well-established design rules, can be programmed to assemble into a wide variety of crystalline architectures (see “Programmable Nanomaterials” subgroup page for more details).
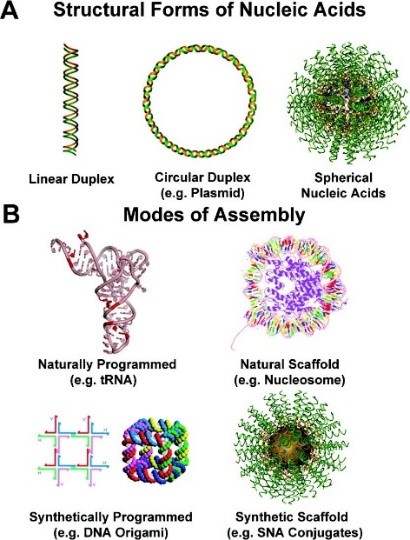
Figure 1. (A) Structural forms of nucleic acids include linear (1D) duplexes (left), circular (2D) plasmid DNA (middle), and spherical (3D) SNAs (right). (B) Natural and synthetic assemblies of nucleic acids. (Taken from J. Am. Chem. Soc. 2012, 134, 3, 1376–1391.)
Video 1. Spherical Nucleic Acids
This animation describes the structure, properties, and uses of SNAs. SNAs have unique architectural-dependent properties that set it apart from all other forms of nucleic acids. These properties make SNAs extremely useful biology and biomedicine.
Structure and Properties of SNA
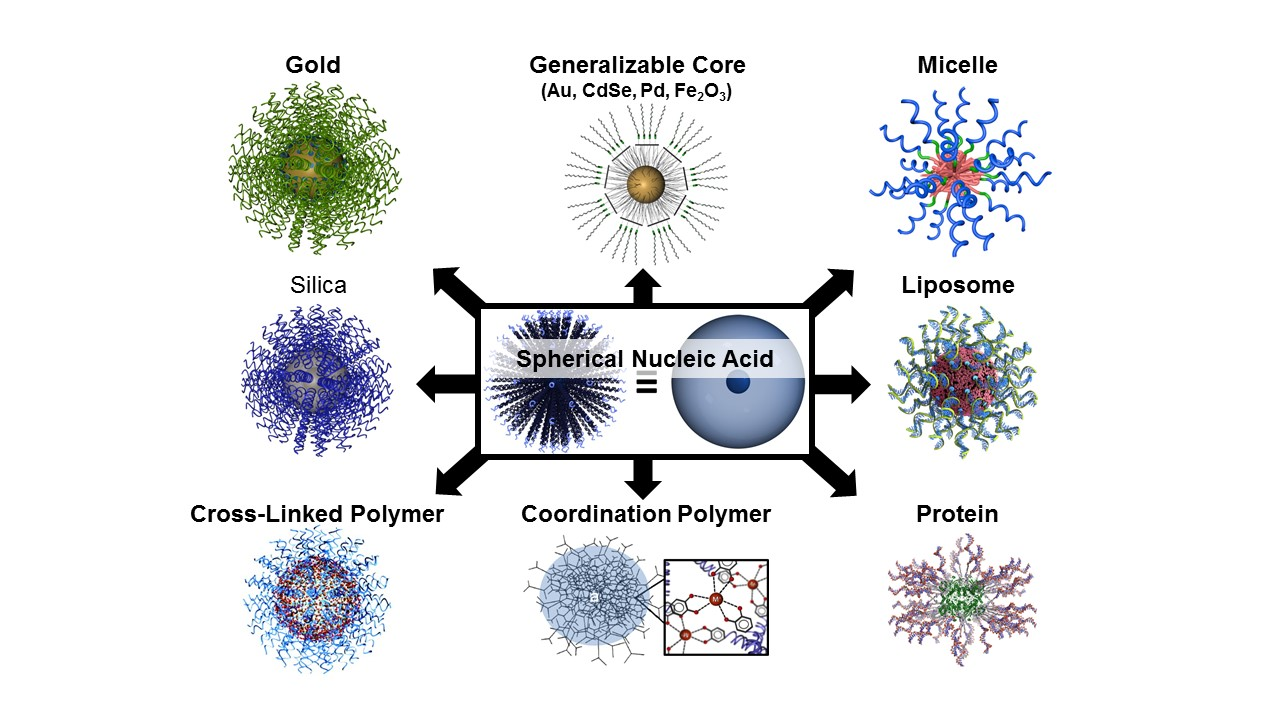
The first form of SNA, invented by the Mirkin group in 1996, consisted of a gold nanoparticle core and a single-stranded DNA (ssDNA) shell. Since then, we and others have developed dozens of SNA structures with different cores (e.g., silica, iron oxide, protein, liposome, micelle, metal organic framework, FDA-approved PLGA, hollow) (Fig. 2) and shells (e.g., DNA, RNA, PNA, LNA). The SNA core serves two purposes: 1) it imparts the conjugate with novel chemical and physical properties, and 2) it acts as a scaffold for assembling and orienting the oligonucleotides into a dense spherical arrangement that gives rise to many of their functional properties. Many of the properties of SNAs stem from the dense layer of oriented nucleic acids and are core-independent. While most forms of nucleic acids rely on the hybridized duplex as the fundamental structural unit that determines their overall shape, SNAs can be prepared from both single- and double-stranded nucleic acids, and their orientation is determined by the shape and composition of the core. SNA nanostructures are distinct from structures made by “DNA origami” – they can be synthesized independent of nucleic acid sequence and hybridization interactions and are typically formed via chemical bonds, rather than recognition processes.
Figure 2. Scheme showing many different types of SNA structures based on different cores and shells.
SNAs as Probes for Biodiagnostic mRNA Detection in Live Cells: The Nanoflare
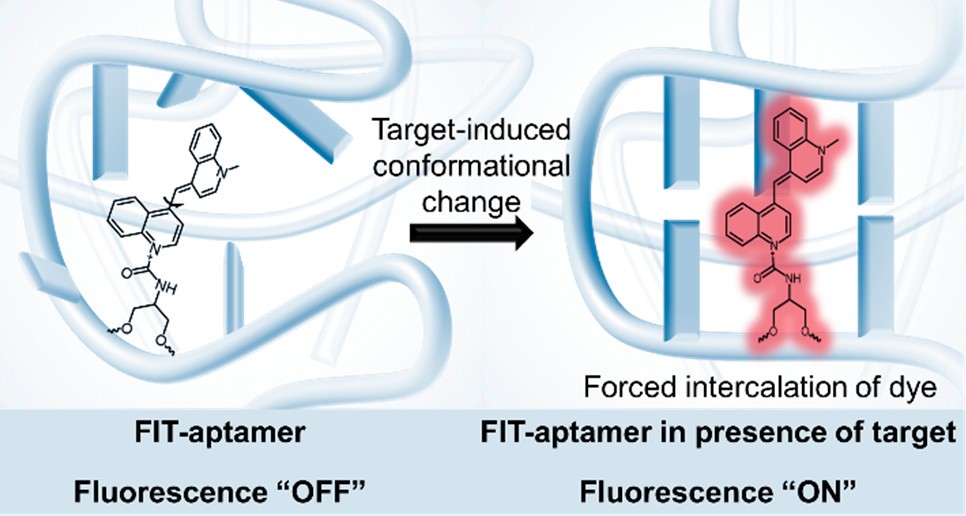
NanoFlares are SNA-based constructs that can be used to detect intracellular targets in live cells at the single-cell level (Fig. 3). The prototypical NanoFlare consists of a spherical gold nanoparticle core densely functionalized with a monolayer of ssDNA. The ssDNA “recognition sequence” is pre-hybridized to a shorter DNA complement containing a fluorescent reporter (the “flare”) whose fluorescence is quenched because of its proximity to the gold particle. When a target binds to the recognition sequence, the flare strand is displaced, providing a fluorescent readout. Nanoflare constructs have been used to detect metastatic markers, such as Vimentin and Twist, and they can be used with whole blood samples to detect circulating tumor cells and diagnose patient risk for metastasis. More recent innovations have led to readout based on forced-intercalation (FIT) (Fig. 4), a “turn on” system with a lower susceptibility to false positives, allowing the researchers to overcome certain limitations of the traditional NanoFlare system. In addition, newer generations of NanoFlares involve the utilization of protein-core systems that employ an i-motif as the surface-based nucleic acid recognition element. These systems allow for the detection of multiple different components because the protein core can act as a functional enzyme. Nanoflares have been commercialized by AuraSense (a company co-founded by Prof. Mirkin) and Merck Millipore as SmartFlares (https://www.emdmillipore.com/US/en/life-science-research/genomic-analysis/SmartFlare-Live-Cell-RNA-Detection/ZdGb.qB.KCcAAAFLAQs0i.s1,nav).
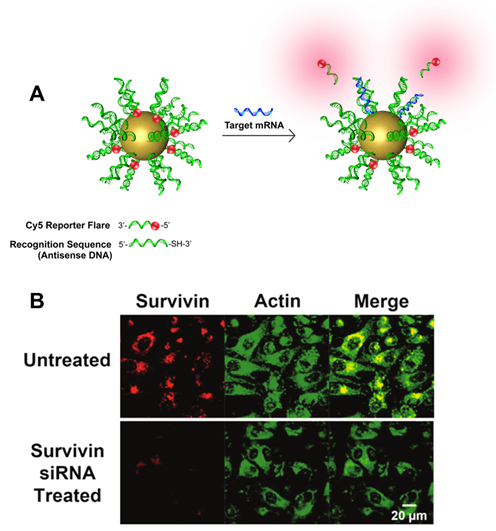
Figure 3. (A) Scheme showing the design and detection process of the NanoFlare. (B) Fluorescence microscope images showing the detection of survivin mRNA in living cells using NanoFlares. (Taken from Anal. Chem., 2012, 84, 2062-2066.)
Figure 4. Schematic for FIT-aptamers. (Taken from J. Am. Chem. Soc., 2019, 141, 35, 13744–13748.)
Ultrasensitive Biomarker Detection: Scanometric Assay
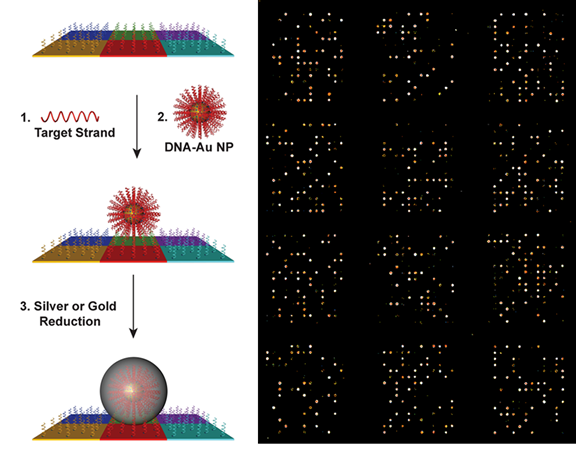
The scanometric assay is a diagnostic tool based on SNA probes that is used for the enzyme-free ultrasensitive detection of various protein and nucleic acid targets. The assay utilizes slides functionalized with biomolecular arrays to capture protein and nucleic acid targets, which are then sandwiched with SNA probes. The signal is amplified by catalytic reduction of Ag(I) or Au(III) in the presence of reducing agents. After the reduction step, the scattered light from the metal spots on the array is used to determine target identity and concentration (Fig. 5). The scanometric assay can be used to detect biomolecules, such as proteins and nucleic acids, at extremely low concentrations. In the case of proteins, such as prostate specific antigen (PSA), the limit of detection (LOD) is orders of magnitude lower than conventional ELISA-based assays. In the case of nucleic acid detection, the LOD is 100 aM for large DNA targets, and the method does not require PCR or related target amplification techniques. The scanometric assay has been also been used to profile the expression of miRNA species from human serum, cell culture, and human tissue samples. The scanometric assay forms the basis for Luminex’s Verigene system (https://www.luminexcorp.com/the-verigene-system/), which was originally commercialized by Nanosphere, a company founded by Prof. Mirkin.
Figure 5. Left: Scheme showing the miRNA profiling process using scanometric technology. (Taken from J. Am. Chem. Soc. 2012, 134, 3, 1376–1391.) Right: Light-scattering images of miRNA profiling pattern on microarray.
SNAs as Therapeutic Agents
SNAs for Gene Regulation
Synthetic oligonucleotides can be used to understand intracellular processes and regulate gene expression, offering a route to viable treatment options for many diseases and disorders with a genetic basis, including many cancers or neurological disorders. However, a major challenge still exists regarding how to deliver synthetic nucleic acids to specific disease sites and into cells. Indeed, natural defense networks exist that work to keep foreign nucleic acids out of cells. Furthermore, nucleic acids are often rapidly degraded by nucleases and can activate an innate immune response. Historically, positively charged transfection agents have been used to shuttle negatively charged nucleic acids through negatively charged cell membranes and protect nucleic acids from enzymatic degradation. Unfortunately, transfection agents have not been ideal for use in systemic delivery because they are not degraded naturally, and they can illicit severe toxicity. SNA constructs provide an alternative in this regard since, despite their high negative charge (zeta potential < −30 mV), they enter cells in very high quantities without the need for ancillary transfection agents, and potently regulate gene expression while resisting nuclease degradation. In addition, they are non-toxic and biocompatible. In one key example, SNAs have been used in the treatment of glioblastoma multiforme (aggressive brain cancer) to target the oncoprotein Bcl2Like12 (Bcl2L12). This protein is overexpressed in GBM relative to normal brain and low-grade astrocytomas. SNAs penetrate the blood-brain barrier (BBB) and blood-tumor barrier (BTB), and they deliver siRNA that knocks down endogenous Bcl2L12 mRNA and protein, ultimately allowing glioma cells to become more sensitive and responsive toward therapy-induced apoptosis.
SNAs for Immunotherapy
Nucleic acids that have been designed to modulate the immune system hold extraordinary promise for treating disease, yet clinical progress has been limited by a lack of tools to safely increase (e.g., in the case of cancers) or decrease (e.g., in the case of auto-immune diseases) immune activity in patients. Certain immunotherapeutic nucleic acids act by agonizing or antagonizing endosomal toll-like receptors (e.g., TLR3, TLR7/8, and TLR9), receptors involved in innate immune signaling. Immunomodulatory SNAs that stimulate (immunostimulatory, IS-SNA) or regulate (immunoregulatory, IR-SNA) immunity by engaging TLRs have been developed by the Mirkin group. Compared to free oligonucleotides, IS-SNAs exhibit up to an 80-fold increase in potency, 700-fold higher antibody titers, 400-fold higher cellular responses to a model antigen, and improved treatment of mice with lymphomas as well as other types of cancers. IR-SNAs exhibit up to eightfold increases in potency and 30% greater reduction in fibrosis score in mice with nonalcoholic steatohepatitis (NASH). More recently, SNAs have been utilized to establish the field of “rational vaccinology,” where the chemical structure of the vaccine has been proven to alter a treatment from being mildly effective to curative (Fig. 6). The Mirkin group is currently working to develop new SNA-based immunotherapies for cancers including triple-negative breast cancer, prostate cancer, and melanoma.
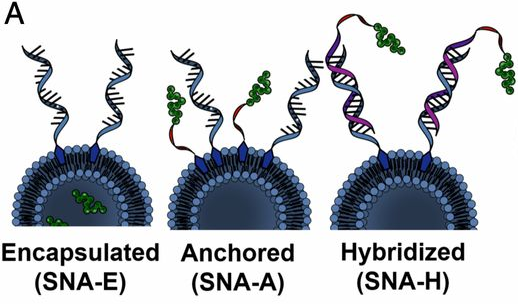
Figure 6. Structures of SNA immunotherapeutics with different antigen and adjuvant arrangements. (Taken from Proc. Natl. Aca. Sci. USA, 2019, 116, 10473-10481.)
SNAs are being commercialized as agents for gene regulation and immunotherapy by Exicure (http://www.exicuretx.com/, Nasdaq: XCUR). SNA drugs have formed the basis of six human clinical trials (two fast-tracked by the FDA) thus far run by Exicure and Northwestern (see Videos 2 and 3), including for the treatment of glioblastoma multiforme and rare skin cancers. One SNA drug, Cavrotolimod (or Cavro), potently stimulates the immune system and holds great promise in cancer treatment and advanced immunotherapeutic development. In September 2020, Exicure presented promising interim results from an ongoing Phase 1b/2 trial of Cavro at a Virtual Key Opinion Leader (KOL) event. Key findings/take-aways include: target tumor shrinkage was observed in 37% of patients; a confirmed overall response rate (ORR) of 21% in the dose-escalation stage across all doses; confirmed ORR 33% at the highest and selected Phase 2 dose. This preliminary data shows activity in patients with melanoma, Merkel cell carcinoma (MCC), and cutaneous squamous cell carcinoma (CSCC). Phase 2 arms in both MCC and CSCC are recruiting.
Videos 2 and 3. Spherical Nucleic Acids in Clinical Trials
Two videos discussing translational efforts with SNAs.
Protein Transfection using SNAs
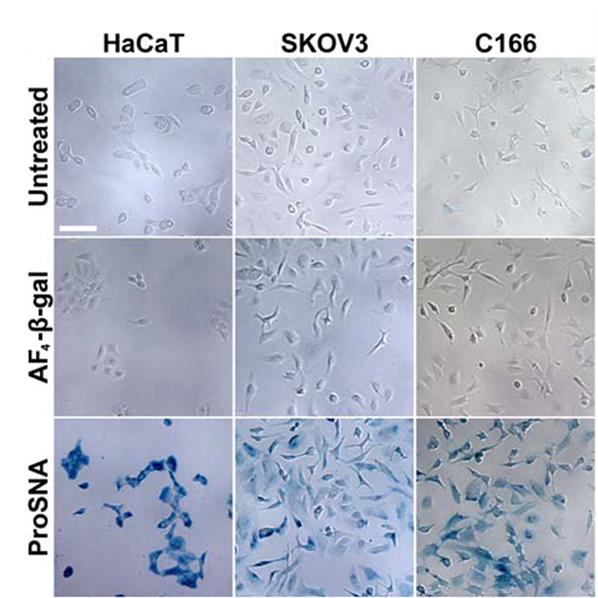
Proteins represent a highly evolved class of natural nanoparticles with an unparalleled degree of structural and compositional homogeneity as well as diversity of functions. In particular, the efficient intracellular delivery of functional proteins has widespread potential in medicine and provides a means for engineering cellular functions. However, the cellular uptake of functional proteins is impeded by their inherent instability, large sizes, and charged surfaces. However, proteins can serve as the core of SNAs, and since it is the nucleic acid shell that is responsible for many of the useful properties of SNA, including enhanced cellular uptake, proteins can be shuttled into cells. This class of protein-core SNAs, termed ProSNAs (Fig. 7), are emerging as a new class of protein transfection materials. β-galactosidase ProSNAs retain the native structure and catalytic ability of the hydrolytic enzyme β-galactosidase, which serves as the protein core, despite the functionalization of its surface with ∼25 DNA strands. The covalent attachment of a shell of oligonucleotides to the surface of β-galactosidase enhances its cellular uptake by up to ∼280-fold and allows for the use of working concentrations as low as 100 pM enzyme. DNA-functionalized β-galactosidase retains its ability to catalyze the hydrolysis of β-glycosidic linkages once endocytosed, whereas equal concentrations of protein show little to no intracellular catalytic activity (Fig. 8). In addition, these ProSNAs retain their enzymatic activity in tissues following systemic injection in vivo.
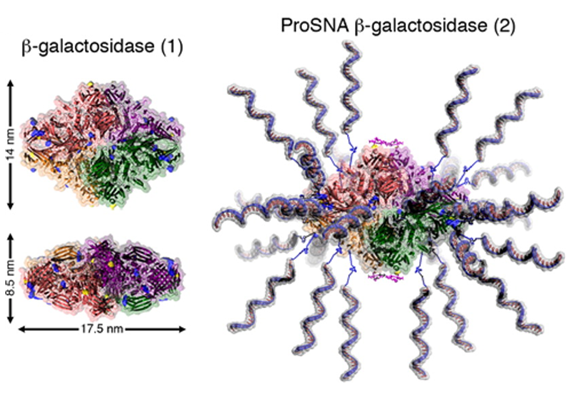
Figure 7. Cartoon representation of β-gal before (left) and after (right) functionalization with DNA. The representation was adapted from PDB ID 1BGL.32. (Taken from J. Am. Chem. Soc. 2015, 137, 14838-14841.)
Figure 8. Intracellular catalytic activity of native and ProSNA β-gal. (A) Light micrographs of HaCaT (left), SKOV3 (middle), and C166 (right) cells after incubation with the β-gal substrate, Xgal. The blue color apparent in cells pretreated with ProSNA β-gal results from the hydrolysis of Xgal and formation of an insoluble reaction product. Scale bar = 100 μm. (Taken from J. Am. Chem. Soc. 2015, 137, 14838-14841.)
Nanomedicine Subgroup Members

Front row (L-R): Yinglun Ma, Alex Cushing, Kathleen Ngo, Janice Kang, Jeongmin Hwang, Julianna Bourgeois (Subgroup Leader), Maya Kesan
Middle row (L-R): Alex Anderson, Jasper Dittmar, John Cavaliere, Nathan Anderson, Tim Kim, Jack Ahrens, Roger Romero, T. Ocampo, Julia Petrescu, Tanner Fink, Mia Pascall
Last row (L-R): Krishna Paranandi, Connor Forsyth, Taokun Luo, Vinzenz Mayer, Jake Cohen, Sergej Kudruk (subgroup Leader)